Jared Homola, Ph.D. student, University of Maine
Understanding how animals move across a landscape is important to the Of Pools and People research team, as well as ecologists studying populations of many other animals. For decades, researchers have used direct tagging methods that physically mark individual animals to learn about their movements. For these approaches, animals are often captured, marked, then must be recaptured to gather data on their movements. In other cases, movement can be tracked by following radio signals or fluorescent tracing powder left behind by the animal. Such tagging studies continue to provide important and reliable information on animal movements; however, these approaches are not ideal for all applications. For instance, repeatedly capturing the same individuals can be difficult, especially for cryptic or rare species. Additionally, data collected from direct tagging usually only provides information on animal movement between capture events. Because of these limitations, direct tagging approaches are sometimes paired with information drawn from the genetics of individuals in order to gain a better understanding of how animals move over extended time and spatial scales.
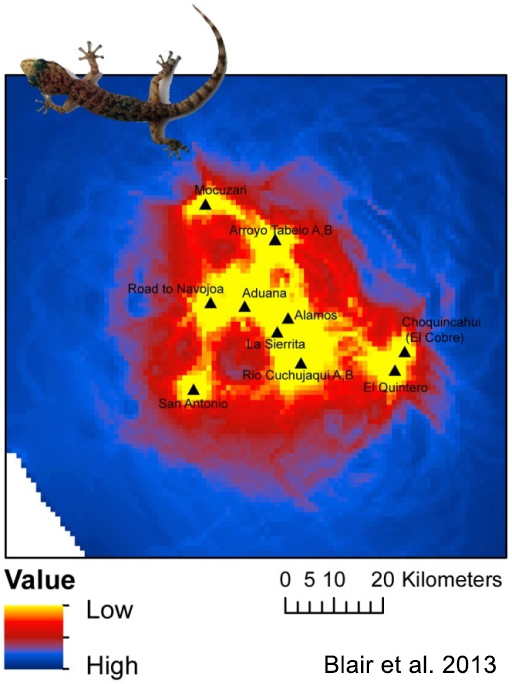
But how can genetics provide information on animal movement? This is often accomplished by comparing how similar the genetics of two (or more) populations are to each other. For example, if two populations are very similar at the genetic level, we can usually assume that individuals are frequently moving between the populations. Conversely, strong differences in genetic characteristics can indicate that individuals are not moving between populations, which could be due to some sort of barrier stopping movement or simply because the populations are spread far from each other on the landscape. In recent years, a wide variety of statistical methods have been developed based on quantifying inter-population genetic differences. These approaches allow researchers to visualize how the landscape can influence movements of animals (see Figure 1), enabling the identification and conservation of habitats that are important to movement.
Because genetic patterns are established over many generations, these approaches provide insight across longer time scales, although at a courser resolution, than direct tagging methods. Additionally, because only a single capture event is necessary to collect tissue for genetic analyses, information can be gathered on species that would otherwise be difficult to track. These unique qualities make genetic analysis of individual movement a valuable complementary approach to more commonly applied direct tagging methods.
References:
Blair, C., V.H.J. Arcos, F.R. Mendez de la Cruz, and R.W. Murphy. 2013. Landscape genetics of leaf-toed geckos in the tropical dry forest of northern Mexico. PLoS One 8(2):e57433.